캐글데이터분석(Mushroom Edible)
Mushroom Edible Classification
Description
Dataset Source :
https://www.kaggle.com/datasets/uciml/mushroom-classification
Problem :
The purpose of this model is to use the categorical properties of mushrooms to classify whether they are edible or not.
- EDA :
CountPlot, Kramer V Coef
- Feature Selection :
Based Kramer V Coef
T1 : Drop gil-attachment, stalk-color-above-ring
Used Features : 19
T2 : Use Odor, spore-print-color
Used Features : 2
- Preprocessing :
LabelEncoder : All Features
OnehotEncoder (unique>=3) ? OnehotEncoder : None
- Modeling :
Train , Test = 8 : 2
All : Stratified K-Fold(N-folds : 5) and Optuna Tuning
Use Model : SVM, DT, MLPClassifier
Hyper Parameter from Optuna, S-kfold(5)
SVM : params {'C': 72.17831419000564, 'kernel': 'poly'}
DT : params {'criterion': 'gini', 'max_depth': 27}
MLPClassifier : params {'learning_rate': 'constant', 'alpha': 0.0001, 'activation': 'tanh'}
- Result :
All Model :
Macro-F1 score : 1.0 , Accuracy : 1.0
Data Load & Simple Analysis
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
data = pd.read_csv('/content/drive/MyDrive/dataset/BIohealthDataset/mushrooms.csv')
data.head(10)
| class | cap-shape | cap-surface | cap-color | bruises | odor | gill-attachment | gill-spacing | gill-size | gill-color | stalk-shape | stalk-root | stalk-surface-above-ring | stalk-surface-below-ring | stalk-color-above-ring | stalk-color-below-ring | veil-type | veil-color | ring-number | ring-type | spore-print-color | population | habitat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | p | x | s | n | t | p | f | c | n | k | e | e | s | s | w | w | p | w | o | p | k | s | u |
| 1 | e | x | s | y | t | a | f | c | b | k | e | c | s | s | w | w | p | w | o | p | n | n | g |
| 2 | e | b | s | w | t | l | f | c | b | n | e | c | s | s | w | w | p | w | o | p | n | n | m |
| 3 | p | x | y | w | t | p | f | c | n | n | e | e | s | s | w | w | p | w | o | p | k | s | u |
| 4 | e | x | s | g | f | n | f | w | b | k | t | e | s | s | w | w | p | w | o | e | n | a | g |
| 5 | e | x | y | y | t | a | f | c | b | n | e | c | s | s | w | w | p | w | o | p | k | n | g |
| 6 | e | b | s | w | t | a | f | c | b | g | e | c | s | s | w | w | p | w | o | p | k | n | m |
| 7 | e | b | y | w | t | l | f | c | b | n | e | c | s | s | w | w | p | w | o | p | n | s | m |
| 8 | p | x | y | w | t | p | f | c | n | p | e | e | s | s | w | w | p | w | o | p | k | v | g |
| 9 | e | b | s | y | t | a | f | c | b | g | e | c | s | s | w | w | p | w | o | p | k | s | m |
결측치는 존재하지 않는다.
data.isnull().sum()
class 0
cap-shape 0
cap-surface 0
cap-color 0
bruises 0
odor 0
gill-attachment 0
gill-spacing 0
gill-size 0
gill-color 0
stalk-shape 0
stalk-root 0
stalk-surface-above-ring 0
stalk-surface-below-ring 0
stalk-color-above-ring 0
stalk-color-below-ring 0
veil-type 0
veil-color 0
ring-number 0
ring-type 0
spore-print-color 0
population 0
habitat 0
dtype: int64
data.describe()
| class | cap-shape | cap-surface | cap-color | bruises | odor | gill-attachment | gill-spacing | gill-size | gill-color | stalk-shape | stalk-root | stalk-surface-above-ring | stalk-surface-below-ring | stalk-color-above-ring | stalk-color-below-ring | veil-type | veil-color | ring-number | ring-type | spore-print-color | population | habitat | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 |
| unique | 2 | 6 | 4 | 10 | 2 | 9 | 2 | 2 | 2 | 12 | 2 | 5 | 4 | 4 | 9 | 9 | 1 | 4 | 3 | 5 | 9 | 6 | 7 |
| top | e | x | y | n | f | n | f | c | b | b | t | b | s | s | w | w | p | w | o | p | w | v | d |
| freq | 4208 | 3656 | 3244 | 2284 | 4748 | 3528 | 7914 | 6812 | 5612 | 1728 | 4608 | 3776 | 5176 | 4936 | 4464 | 4384 | 8124 | 7924 | 7488 | 3968 | 2388 | 4040 | 3148 |
속성 정보: (등급: 식용=e, 독성=p)
모자 모양: 벨=b, 원추형=c, 볼록=x, 평면=f, 손잡이=k,가라앉은=s
캡 표면: 섬유질=f,홈=g,비늘 모양=y,매끄러운=s
캡 색상: 갈색=n,담황색=b,시나몬=c,회색=g,녹색=r,분홍색=p,보라색=u,빨간색=e,흰색=w,노란색=y
타박상: 타박상=t,아니오=f
냄새: 아몬드=a,아니스=l,크레오소트=c,비린내=y,파울=f,곰팡이=m,없음=n,매운=p,매운=s
아가미 부착: 부착=a,내림차순=d,자유=f,노치=n
아가미 간격: 닫기=c, 붐비는=w, 먼=d
아가미 크기: 넓음=b, 좁음=n
아가미 색상: 검정=k,갈색=n,버프=b,초콜릿=h,회색=g, 녹색=r,주황색=o,분홍색=p,보라색=u,빨간색=e,흰색=w,노란색= 와이
줄기 모양: 확대=e, 테이퍼링=t
줄기-뿌리: 구근=b,클럽=c,컵=u,동일=e,근근형=z,뿌리=r,누락=?
줄기-표면-위-고리: 섬유질=f,비늘=y,실키=k,매끄러운=s
줄기-표면-아래-고리: 섬유질=f,비늘=y,실키=k,부드러운=s
고리 위의 줄기 색상: 갈색=n,담황색=b,시나몬=c,회색=g, 주황색=o,분홍색=p,빨간색=e,흰색=w,노란색=y
줄기색-아래-고리: 갈색=n,담황색=b,시나몬=c,회색=g,주황색=o,분홍색=p,빨간색=e,흰색=w,노란색=y
베일형: 부분=p,보편=u
베일 색상: 갈색=n,주황색=o,흰색=w,노란색=y
링 번호: 없음=n, 1=o, 2=t
반지 유형: cobwebby=c,evanescent=e,flaring=f,large=l,none=n,pendant=p,sheathing=s,zone=z
spore-print-color: 검정=k,갈색=n,버프=b,초콜릿=h,녹색=r,주황색=o,보라색=u,흰색=w,노란색=y
인구: 풍부=a,군집=c,수많은=n,흩어진=s,여러=v,단독=y
서식지: 잔디=g,잎=l,초원=m,경로=p,도시=u,폐기물=w,나무=d
data.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 8124 entries, 0 to 8123
Data columns (total 23 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 class 8124 non-null object
1 cap-shape 8124 non-null object
2 cap-surface 8124 non-null object
3 cap-color 8124 non-null object
4 bruises 8124 non-null object
5 odor 8124 non-null object
6 gill-attachment 8124 non-null object
7 gill-spacing 8124 non-null object
8 gill-size 8124 non-null object
9 gill-color 8124 non-null object
10 stalk-shape 8124 non-null object
11 stalk-root 8124 non-null object
12 stalk-surface-above-ring 8124 non-null object
13 stalk-surface-below-ring 8124 non-null object
14 stalk-color-above-ring 8124 non-null object
15 stalk-color-below-ring 8124 non-null object
16 veil-type 8124 non-null object
17 veil-color 8124 non-null object
18 ring-number 8124 non-null object
19 ring-type 8124 non-null object
20 spore-print-color 8124 non-null object
21 population 8124 non-null object
22 habitat 8124 non-null object
dtypes: object(23)
memory usage: 1.4+ MB
Unique한 값
for i in data.columns:
print(i, " : " ,data[i].nunique())
class : 2
cap-shape : 6
cap-surface : 4
cap-color : 10
bruises : 2
odor : 9
gill-attachment : 2
gill-spacing : 2
gill-size : 2
gill-color : 12
stalk-shape : 2
stalk-root : 5
stalk-surface-above-ring : 4
stalk-surface-below-ring : 4
stalk-color-above-ring : 9
stalk-color-below-ring : 9
veil-type : 1
veil-color : 4
ring-number : 3
ring-type : 5
spore-print-color : 9
population : 6
habitat : 7
EDA
data.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 8124 entries, 0 to 8123
Data columns (total 23 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 class 8124 non-null object
1 cap-shape 8124 non-null object
2 cap-surface 8124 non-null object
3 cap-color 8124 non-null object
4 bruises 8124 non-null object
5 odor 8124 non-null object
6 gill-attachment 8124 non-null object
7 gill-spacing 8124 non-null object
8 gill-size 8124 non-null object
9 gill-color 8124 non-null object
10 stalk-shape 8124 non-null object
11 stalk-root 8124 non-null object
12 stalk-surface-above-ring 8124 non-null object
13 stalk-surface-below-ring 8124 non-null object
14 stalk-color-above-ring 8124 non-null object
15 stalk-color-below-ring 8124 non-null object
16 veil-type 8124 non-null object
17 veil-color 8124 non-null object
18 ring-number 8124 non-null object
19 ring-type 8124 non-null object
20 spore-print-color 8124 non-null object
21 population 8124 non-null object
22 habitat 8124 non-null object
dtypes: object(23)
memory usage: 1.4+ MB
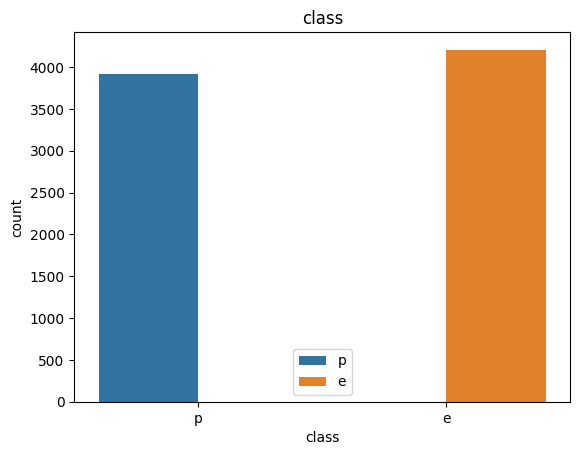
data['class'].value_counts()
e 4208
p 3916
Name: class, dtype: int64
def count_plot(data, col):
sns.countplot(data=data,x=data[col],hue=data['class'])
plt.title(col)
plt.legend()
plt.show()
for col in data.columns:
count_plot(data,col)
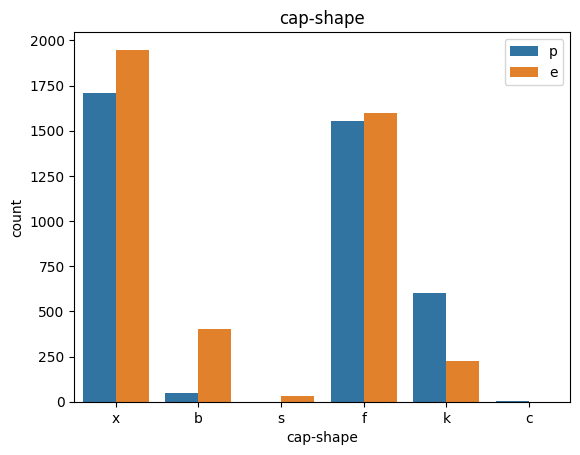
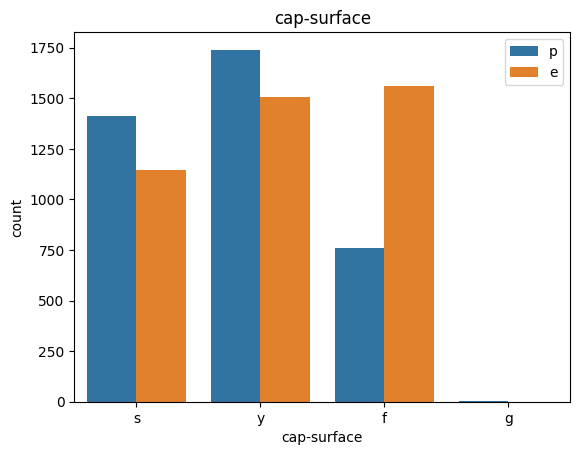
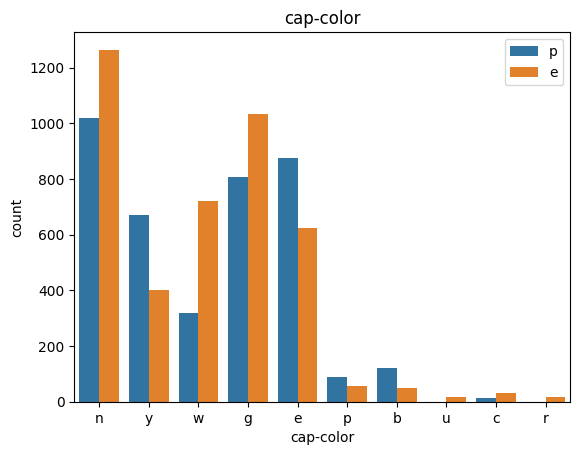
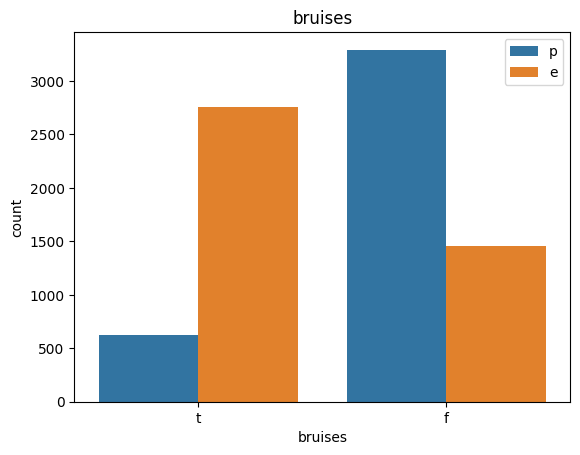
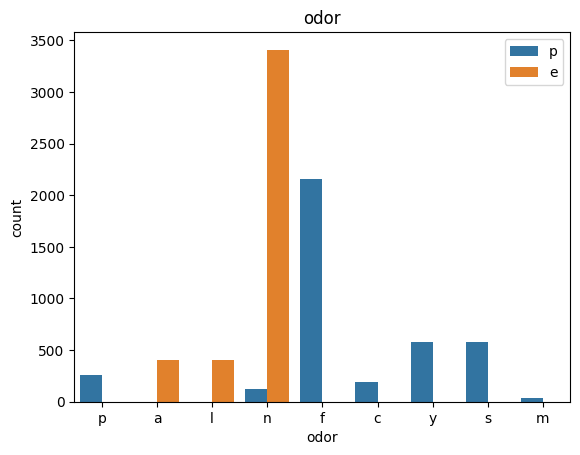
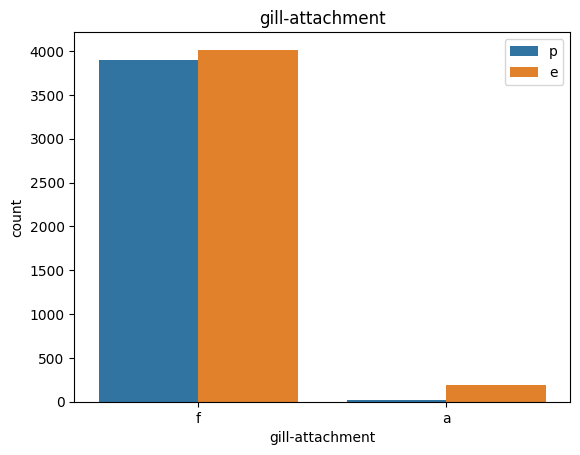
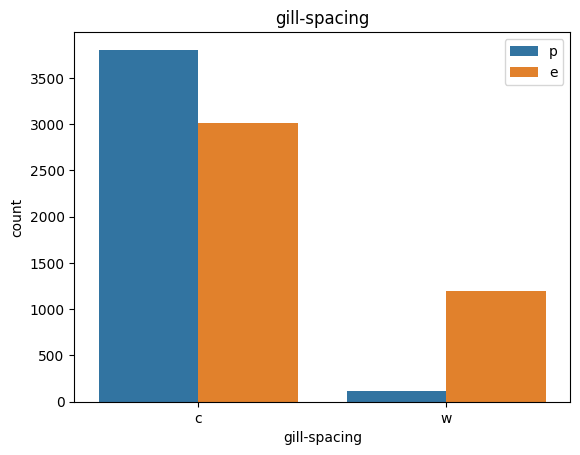
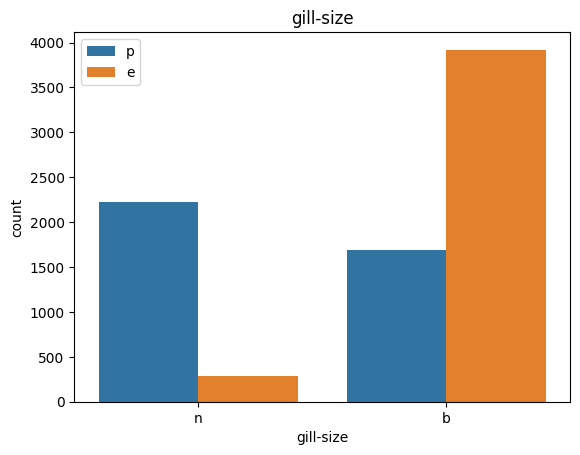
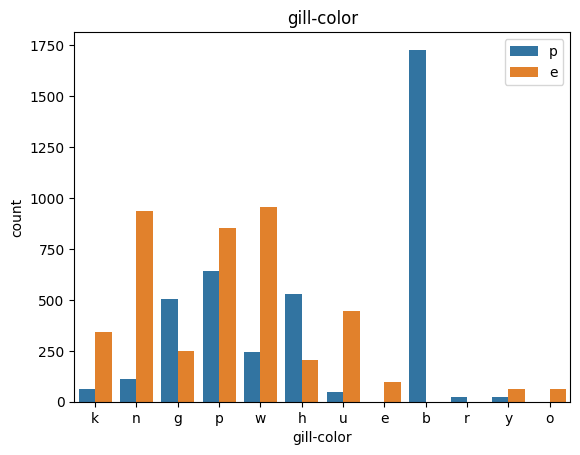
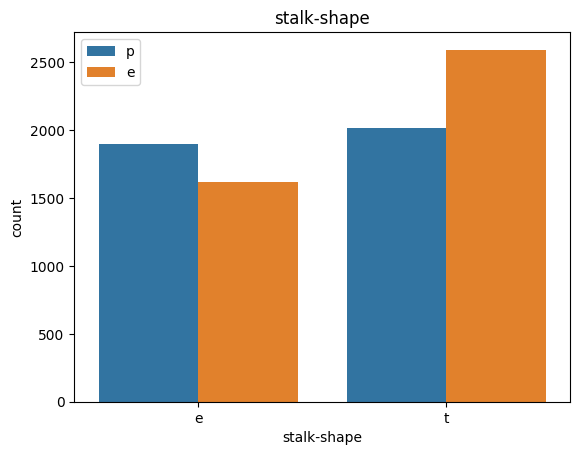
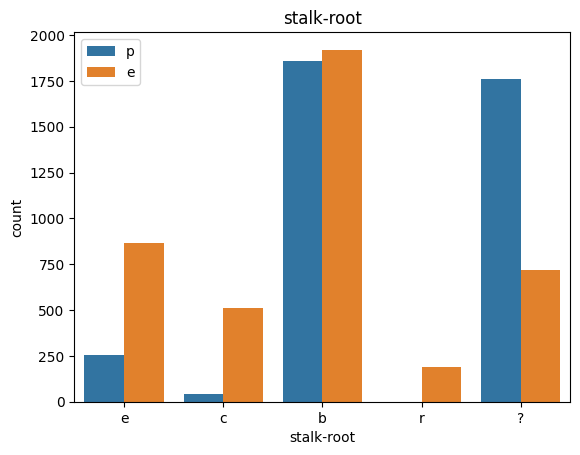
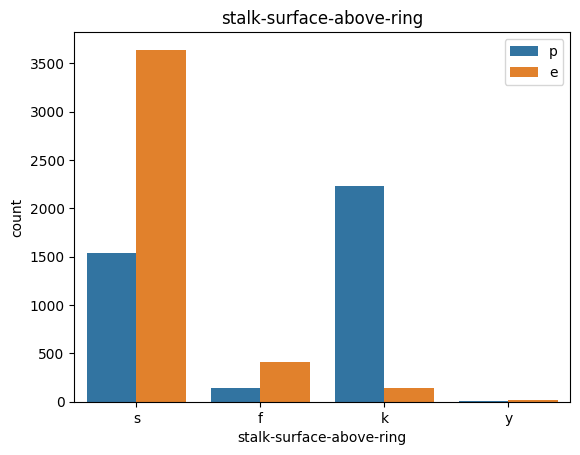
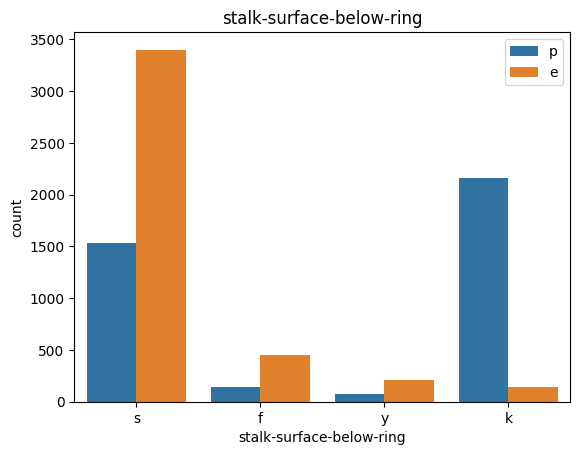
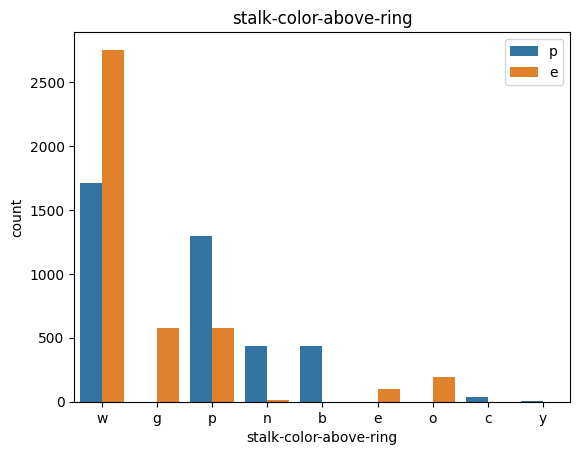
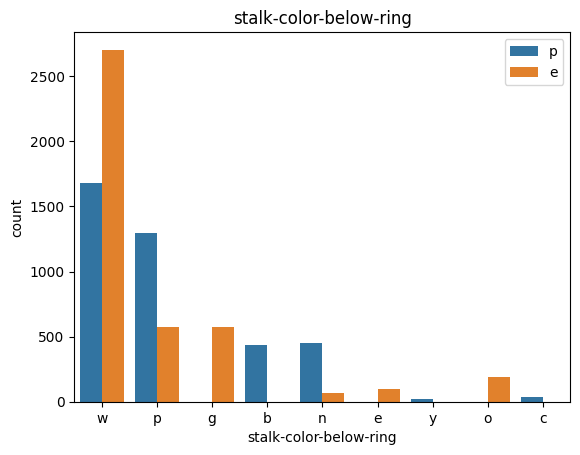
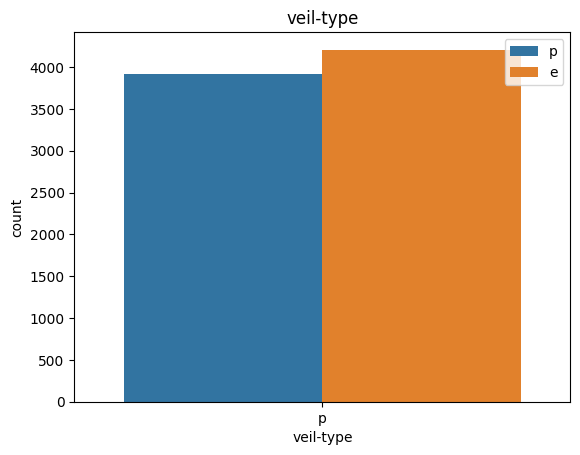
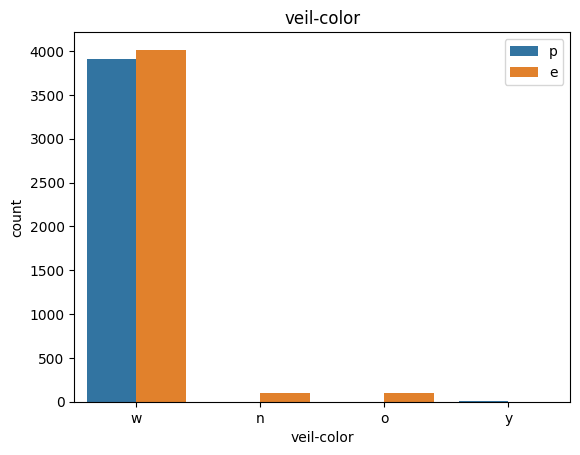
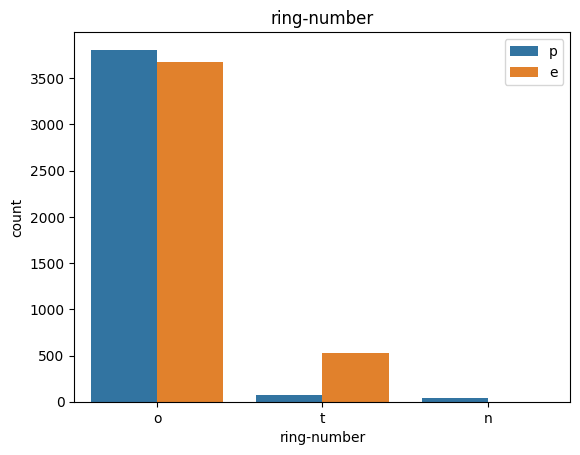
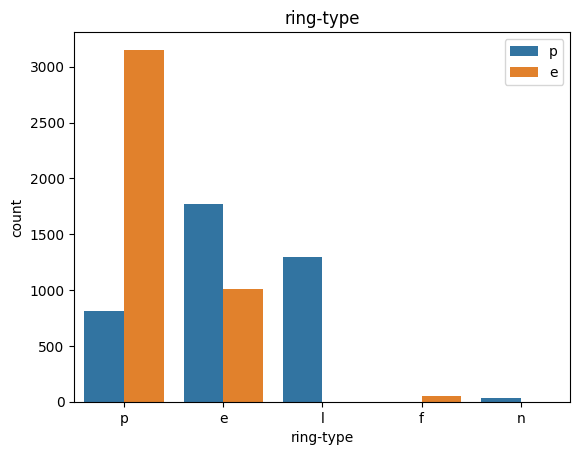
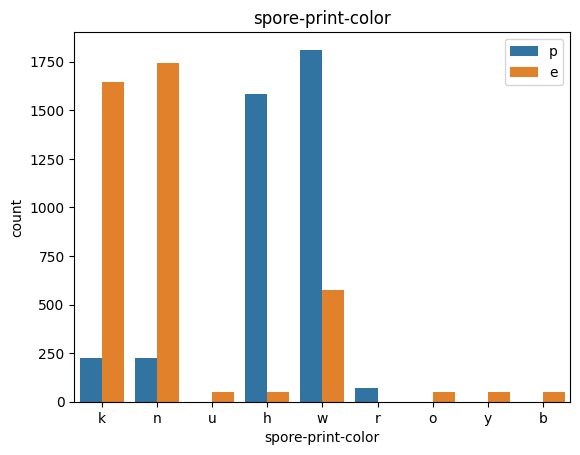
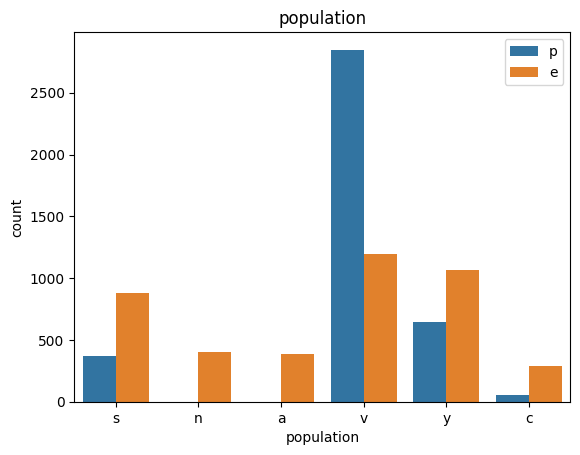
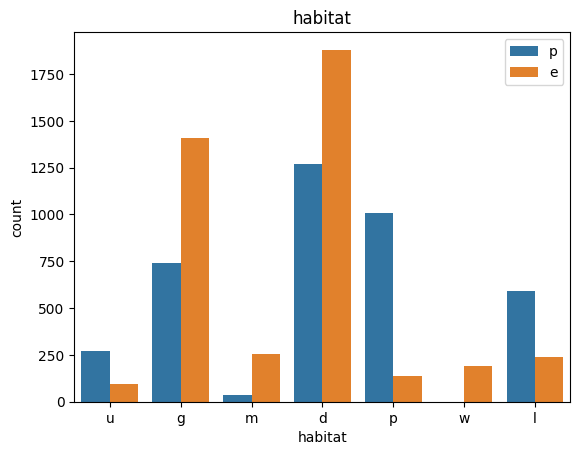
# veil특성은 unique값이 1이므로, 제외한다.
data =data.drop('veil-type',axis=1)
크라머 상관계수 분석
from sklearn.feature_selection import chi2, RFECV
from sklearn.model_selection import train_test_split, cross_validate, GridSearchCV
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC, LinearSVC
from sklearn.tree import DecisionTreeClassifier, plot_tree
import scipy.stats as ss
from sklearn.model_selection import train_test_split
train_df, test_df=train_test_split(data,test_size=.2,random_state=5)
train_df['class'].value_counts()
e 3381
p 3118
Name: class, dtype: int64
test_df['class'].value_counts()
e 827
p 798
Name: class, dtype: int64
train_df.columns
Index(['class', 'cap-shape', 'cap-surface', 'cap-color', 'bruises', 'odor',
'gill-attachment', 'gill-spacing', 'gill-size', 'gill-color',
'stalk-shape', 'stalk-root', 'stalk-surface-above-ring',
'stalk-surface-below-ring', 'stalk-color-above-ring',
'stalk-color-below-ring', 'veil-color', 'ring-number', 'ring-type',
'spore-print-color', 'population', 'habitat'],
dtype='object')
def cramers_v(confusion_matrix):
chi2 = ss.chi2_contingency(confusion_matrix)[0]
n = confusion_matrix.sum()
phi2 = chi2 / n
r, k = confusion_matrix.shape
phi2corr = max(0, phi2 - ((k-1)*(r-1))/(n-1))
rcorr = r - ((r-1)**2)/(n-1)
kcorr = k - ((k-1)**2)/(n-1)
return np.sqrt(phi2corr / min((kcorr-1), (rcorr-1)))
rows= []
for var1 in data:
col = []
for var2 in data :
confusion_matrix = pd.crosstab(data[var1], data[var2])
# Cramer's V test
col.append(round(cramers_v(confusion_matrix.values),2)) # Keeping of the rounded value of the Cramer's V
rows.append(col)
cramers_results = np.array(rows)
df_corr = pd.DataFrame(cramers_results, columns = data.columns, index =data.columns)
plt.figure(figsize=(30,15))
sns.heatmap(df_corr[df_corr > 0.6], annot=True, cmap=plt.cm.CMRmap_r)
plt.show()
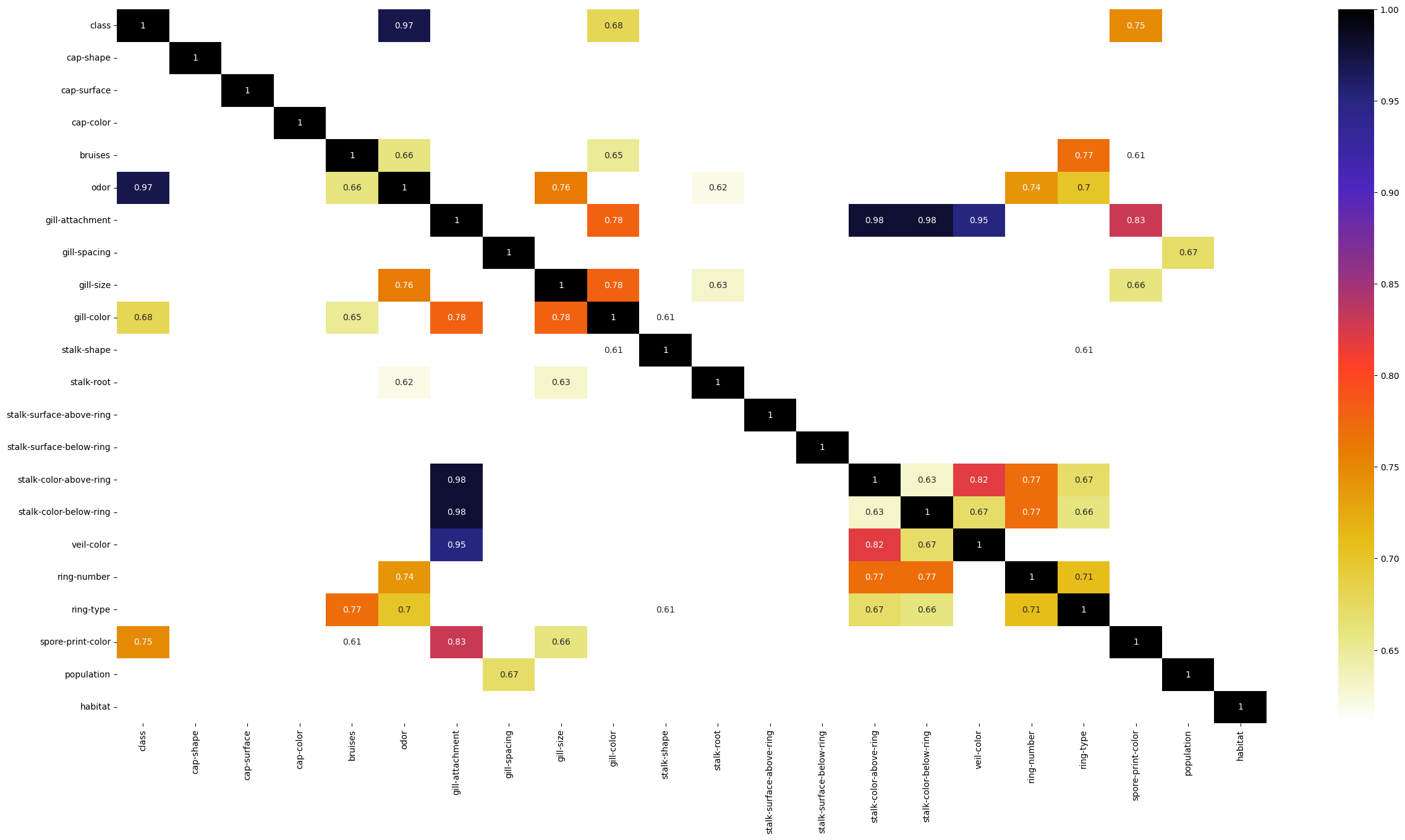
TRY1 : gil-attachment, stalk-color-above-ring을 삭제해주고 진행하겠다.
train_col=list(train_df.columns)
train_col.remove('gill-attachment')
train_col.remove('stalk-color-above-ring')
TRY2 : Target과 연관성이 높은 odor , spore-print-color만 사용하겠다.
train_col = ['odor','spore-print-color','class']
train_col
['class',
'cap-shape',
'cap-surface',
'cap-color',
'bruises',
'odor',
'gill-spacing',
'gill-size',
'gill-color',
'stalk-shape',
'stalk-root',
'stalk-surface-above-ring',
'stalk-surface-below-ring',
'stalk-color-below-ring',
'veil-color',
'ring-number',
'ring-type',
'spore-print-color',
'population',
'habitat']
Data Preprocessing
모든 컬럼들이 Categorical하기때문에, unique한값이 3이상인 값들에 대해서 원핫인코딩 진행하고, unique 값이 2인 컬럼들은 라벨인코더만 진행한다.
train_df=train_df[train_col]
test_df = test_df[train_col]
one_hot_cols=list(train_df.columns[train_df.nunique()>2])
binary_cols=list(train_df.columns[train_df.nunique()==2])
binary_cols
['class']
one_hot_cols
['odor', 'spore-print-color']
unique한 값이 3이상인 값들에 대해서만 원핫인코딩을 진행한다. 2인값들에 대해서는 라벨인코더만 진행한다.
일단 전체 column에 대해 LabelEncoder작업을 진행해준다.
train_df.nunique()
class 2
stalk-shape 2
dtype: int64
from sklearn.preprocessing import LabelEncoder
enc = LabelEncoder()
for col in train_df.columns:
enc.fit(train_df[col])
train_df[col]=enc.transform(train_df[col])
# Data Leakage에 유의한다.
test_df[col]=enc.transform(test_df[col])
train_df
| class | stalk-shape | |
|---|---|---|
| 5174 | 1 | 0 |
| 7927 | 0 | 0 |
| 2641 | 0 | 1 |
| 5669 | 0 | 0 |
| 2043 | 0 | 1 |
| ... | ... | ... |
| 3046 | 0 | 1 |
| 1725 | 1 | 0 |
| 4079 | 1 | 0 |
| 2254 | 0 | 1 |
| 2915 | 0 | 1 |
6499 rows × 2 columns
test_df
| class | gill-color | |
|---|---|---|
| 5350 | 1 | 3 |
| 7926 | 1 | 0 |
| 4804 | 1 | 2 |
| 569 | 0 | 10 |
| 1844 | 0 | 7 |
| ... | ... | ... |
| 529 | 0 | 10 |
| 6348 | 1 | 0 |
| 4659 | 1 | 2 |
| 5700 | 1 | 7 |
| 4578 | 1 | 3 |
1625 rows × 2 columns
인덱스 초기화
train_df = train_df.reset_index(drop=True)
test_df = test_df.reset_index(drop=True)
One hot Encoding
from sklearn.preprocessing import OneHotEncoder
ohe = OneHotEncoder(sparse=False)
# fit_transform은 train에만 사용하고 test에는 학습된 인코더에 transform만 해야한다
for col in one_hot_cols:
ohe.fit(train_df[[col]])
train_cat=ohe.transform(train_df[[col]])
train_df=pd.concat([train_df.drop(columns=[col]),
pd.DataFrame(train_cat, columns=[col+"_"+ str(c) for c in ohe.categories_[0]])], axis=1)
test_cat = ohe.transform(test_df[[col]])
test_df = pd.concat([test_df.drop(columns=[col]),
pd.DataFrame(test_cat, columns=[ col +"_"+ str(c) for c in ohe.categories_[0]])], axis=1)
/usr/local/lib/python3.9/dist-packages/sklearn/preprocessing/_encoders.py:868: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
/usr/local/lib/python3.9/dist-packages/sklearn/preprocessing/_encoders.py:868: FutureWarning: `sparse` was renamed to `sparse_output` in version 1.2 and will be removed in 1.4. `sparse_output` is ignored unless you leave `sparse` to its default value.
warnings.warn(
test_df
| class | stalk-shape | |
|---|---|---|
| 0 | 1 | 1 |
| 1 | 1 | 1 |
| 2 | 1 | 0 |
| 3 | 0 | 0 |
| 4 | 0 | 1 |
| ... | ... | ... |
| 1620 | 0 | 0 |
| 1621 | 1 | 1 |
| 1622 | 1 | 0 |
| 1623 | 1 | 0 |
| 1624 | 1 | 0 |
1625 rows × 2 columns
y=train_df['class']
train=train_df.drop('class',axis=1)
y_test = test_df['class']
test=test_df.drop('class',axis=1)
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.neural_network import MLPClassifier
from sklearn.metrics import accuracy_score, confusion_matrix, f1_score, auc, roc_curve, ConfusionMatrixDisplay
Modeling
SVC
DT
NN
Confusion Matrix 출력을 위한 함수를 미리 생성한다.
def show_confusion(pred,y,model_name):
confusionmatrix = confusion_matrix(y, pred)
cm_display = ConfusionMatrixDisplay(confusionmatrix)
cm_display.plot()
plt.title(model_name)
plt.show()
SVC
Optuna를 통해서 valid-set에 대해서 최적 하이퍼 파라미터를 추출하고, 이를 test set에 적용시킨다.
pip install optuna
Looking in indexes: https://pypi.org/simple, https://us-python.pkg.dev/colab-wheels/public/simple/
Collecting optuna
Downloading optuna-3.1.1-py3-none-any.whl (365 kB)
[2K [90m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━[0m [32m365.7/365.7 kB[0m [31m11.4 MB/s[0m eta [36m0:00:00[0m
[?25hRequirement already satisfied: numpy in /usr/local/lib/python3.9/dist-packages (from optuna) (1.22.4)
Collecting cmaes>=0.9.1
Downloading cmaes-0.9.1-py3-none-any.whl (21 kB)
Requirement already satisfied: packaging>=20.0 in /usr/local/lib/python3.9/dist-packages (from optuna) (23.0)
Requirement already satisfied: tqdm in /usr/local/lib/python3.9/dist-packages (from optuna) (4.65.0)
Requirement already satisfied: sqlalchemy>=1.3.0 in /usr/local/lib/python3.9/dist-packages (from optuna) (2.0.9)
Collecting colorlog
Downloading colorlog-6.7.0-py2.py3-none-any.whl (11 kB)
Requirement already satisfied: PyYAML in /usr/local/lib/python3.9/dist-packages (from optuna) (6.0)
Collecting alembic>=1.5.0
Downloading alembic-1.10.3-py3-none-any.whl (212 kB)
[2K [90m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━[0m [32m212.3/212.3 kB[0m [31m25.9 MB/s[0m eta [36m0:00:00[0m
[?25hCollecting Mako
Downloading Mako-1.2.4-py3-none-any.whl (78 kB)
[2K [90m━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━[0m [32m78.7/78.7 kB[0m [31m10.2 MB/s[0m eta [36m0:00:00[0m
[?25hRequirement already satisfied: typing-extensions>=4 in /usr/local/lib/python3.9/dist-packages (from alembic>=1.5.0->optuna) (4.5.0)
Requirement already satisfied: greenlet!=0.4.17 in /usr/local/lib/python3.9/dist-packages (from sqlalchemy>=1.3.0->optuna) (2.0.2)
Requirement already satisfied: MarkupSafe>=0.9.2 in /usr/local/lib/python3.9/dist-packages (from Mako->alembic>=1.5.0->optuna) (2.1.2)
Installing collected packages: Mako, colorlog, cmaes, alembic, optuna
Successfully installed Mako-1.2.4 alembic-1.10.3 cmaes-0.9.1 colorlog-6.7.0 optuna-3.1.1
import optuna
from optuna import Trial
from optuna.samplers import TPESampler
from sklearn.model_selection import StratifiedKFold
# 데이터 분리에서 x_train , x_valid , y_train , y_valid
#분리해놓은 것을 통해 최적 파라미터를 탐색한다.
sampler = TPESampler(seed=10)
# define function
def objective(trial):
cbrm_param = {
"C" : trial.suggest_float('C', 0.01, 100.0,),
"kernel" : trial.suggest_categorical("kernel", ["linear", "poly", "rbf"]),
"probability" : True
}
# Generate model
model_cbrm =SVC(**cbrm_param )
# k겹 교차검증 실시
n_folds=5
kf = StratifiedKFold(n_splits=n_folds, shuffle=True)
# 추후 메타 모델이 사용할 학습 데이터 반환을 위한 넘파이 배열 초기화
m_f1 =0.0
for folder_counter, (train_index, valid_index) in enumerate(kf.split(train,y)):
# 입력된 학습 데이터에서 기반 모델이 학습/예측할 폴드 데이터 세트 추출
print('\t 폴드 세트: ',folder_counter+1,' 시작')
X_tr = train.loc[train_index]
y_tr = y.loc[train_index]
X_te = train.loc[valid_index]
y_te = y.loc[valid_index]
model_cbrm = model_cbrm.fit(X_tr, y_tr)
pred=model_cbrm.predict(X_te)
print("macro-f1 score : ",f1_score(y_te,pred,average='macro'))
m_f1+=f1_score(y_te,pred,average='macro')
# 평가지표 원하는 평가 지표가 있을 시 바꾸어 준다.
m_f1 = m_f1 / n_folds
return m_f1
hold_model=SVC()
hold_model.fit(train,y)
test_pred=hold_model.predict(test)
accuracy_score(y_test,test_pred)
0.9938461538461538
optuna_cbrm = optuna.create_study(direction='maximize', sampler=sampler)
optuna_cbrm.optimize(objective, n_trials=3)
[32m[I 2023-04-13 13:53:14,435][0m A new study created in memory with name: no-name-df3fec2e-ba0a-43b9-9edd-10bfd90e1988[0m
폴드 세트: 1 시작
macro-f1 score : 0.9884308082237375
폴드 세트: 2 시작
macro-f1 score : 0.9961464377374016
폴드 세트: 3 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 4 시작
[32m[I 2023-04-13 13:53:15,423][0m Trial 0 finished with value: 0.9941399574362515 and parameters: {'C': 0.40478715012816596, 'kernel': 'poly'}. Best is trial 0 with value: 0.9941399574362515.[0m
macro-f1 score : 0.9946035027418356
폴드 세트: 5 시작
macro-f1 score : 0.9969147677881807
폴드 세트: 1 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 2 시작
macro-f1 score : 0.9915185426602477
폴드 세트: 3 시작
macro-f1 score : 0.9938330170777988
폴드 세트: 4 시작
macro-f1 score : 0.9953747628083491
폴드 세트: 5 시작
[32m[I 2023-04-13 13:53:16,066][0m Trial 1 finished with value: 0.9941404214645356 and parameters: {'C': 72.17831419000564, 'kernel': 'poly'}. Best is trial 1 with value: 0.9941404214645356.[0m
macro-f1 score : 0.9953715140861799
폴드 세트: 1 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 2 시작
macro-f1 score : 0.9930616463823603
폴드 세트: 3 시작
macro-f1 score : 0.9922901558337425
폴드 세트: 4 시작
[32m[I 2023-04-13 13:53:16,892][0m Trial 2 finished with value: 0.9941403386299712 and parameters: {'C': 54.259011357446006, 'kernel': 'rbf'}. Best is trial 1 with value: 0.9941404214645356.[0m
macro-f1 score : 0.9953747628083491
폴드 세트: 5 시작
macro-f1 score : 0.9945997181377133
cbrm_trial = optuna_cbrm.best_trial
cbrm_trial_params = cbrm_trial.params
print('Best Trial: score {},\nparams {}'.format(cbrm_trial.value, cbrm_trial_params))
Best Trial: score 0.9941404214645356,
params {'C': 72.17831419000564, 'kernel': 'poly'}
model = SVC(**cbrm_trial_params,probability=True)
model.fit(train_df,y)
SVC(C=72.17831419000564, kernel='poly', probability=True)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
SVC(C=72.17831419000564, kernel='poly', probability=True)
y_test_pred=model.predict(test_df)
y_test_proba=model.predict_proba(test_df)
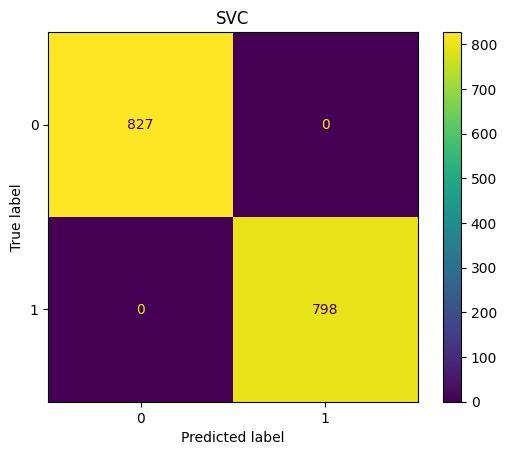
accuracy_score(y_test,y_test_pred)
1.0
f1_score(y_test,y_test_pred,average="macro")
1.0
SVC Confusion Matrix
show_confusion(y_test_pred,y_test,"SVC")
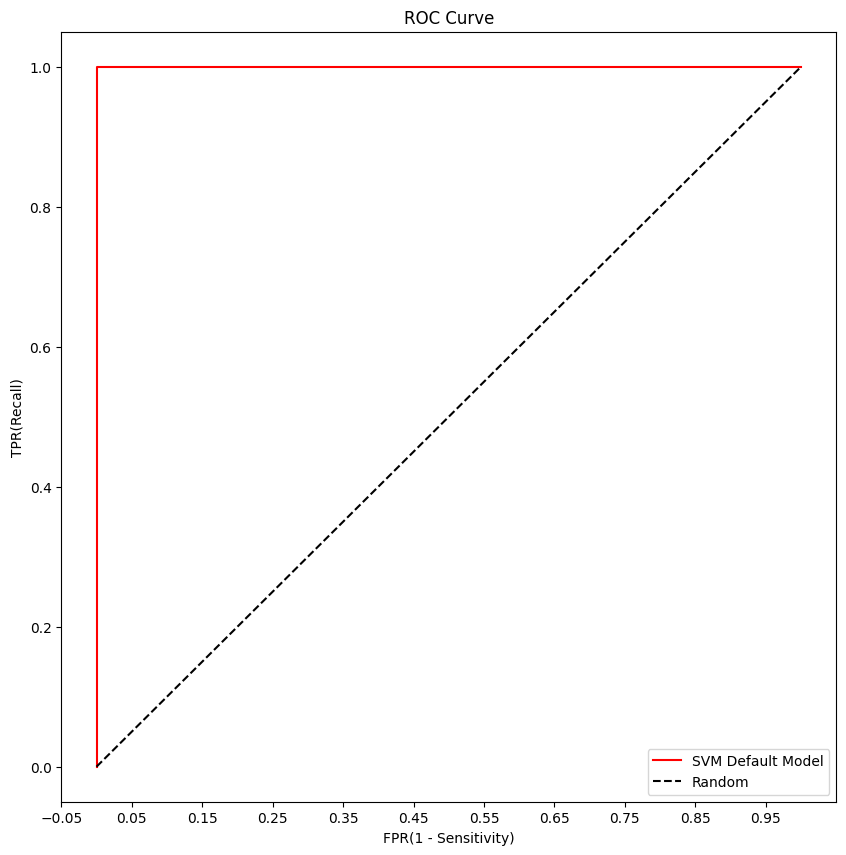
SVC Roc Curve
plt.figure(figsize=(10,10))
fprs, tprs, thresholds = roc_curve(y_test, y_test_proba[:,1])
# ROC 곡선 그래프 곡선으로 그림
plt.plot(fprs, tprs, label='SVM Default Model',color='red')
plt.plot([0,1],[0,1], 'k--', label='Random')
# FPR X 축의 Scale을 0.1 단위로 변경, X, Y 축 명 설정 등
start,end = plt.xlim()
plt.xticks(np.round(np.arange(start, end, 0.1), 2))
#plt.xlim(0, 1); plt.ylim(0, 1)
plt.xlabel('FPR(1 - Sensitivity)'); plt.ylabel('TPR(Recall)')
plt.title('ROC Curve')
plt.legend()
<matplotlib.legend.Legend at 0x7f3f341900a0>
DecisionTree
파라미터 튜닝
# 데이터 분리에서 x_train , x_valid , y_train , y_valid
#분리해놓은 것을 통해 최적 파라미터를 탐색한다.
sampler = TPESampler(seed=10)
# define function
def objective(trial):
cbrm_param = {
"criterion" : trial.suggest_categorical("criterion", ["gini", "entropy","log_loss"]),
"max_depth" : trial.suggest_int("max_depth",4,30)
}
# Generate model
n_folds=5
kf = StratifiedKFold(n_splits=n_folds, shuffle=True)
# 추후 메타 모델이 사용할 학습 데이터 반환을 위한 넘파이 배열 초기화
model_cbrm =DecisionTreeClassifier(**cbrm_param )
m_f1 =0.0
for folder_counter, (train_index, valid_index) in enumerate(kf.split(train,y)):
# 입력된 학습 데이터에서 기반 모델이 학습/예측할 폴드 데이터 세트 추출
print('\t 폴드 세트: ',folder_counter+1,' 시작')
X_tr = train.loc[train_index]
y_tr = y.loc[train_index]
X_te = train.loc[valid_index]
y_te = y.loc[valid_index]
model_cbrm = model_cbrm.fit(X_tr, y_tr)
pred=model_cbrm.predict(X_te)
print("macro-f1 score : ",f1_score(y_te,pred,average='macro'))
m_f1+=f1_score(y_te,pred,average='macro')
# 평가지표 원하는 평가 지표가 있을 시 바꾸어 준다.
m_f1 = m_f1 / n_folds
return m_f1
optuna_cbrm = optuna.create_study(direction='maximize', sampler=sampler)
optuna_cbrm.optimize(objective, n_trials=20)
[32m[I 2023-04-13 13:54:07,810][0m A new study created in memory with name: no-name-538ad95e-3e47-47e0-867c-25aa774359f3[0m
폴드 세트: 1 시작
macro-f1 score : 0.9930616463823603
폴드 세트: 2 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 3 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 4 시작
macro-f1 score : 0.993832124666994
폴드 세트: 5 시작
[32m[I 2023-04-13 13:54:08,066][0m Trial 0 finished with value: 0.9941404061134543 and parameters: {'criterion': 'gini', 'max_depth': 24}. Best is trial 0 with value: 0.9941404061134543.[0m
macro-f1 score : 0.9945997181377133
폴드 세트: 1 시작
macro-f1 score : 0.9922901558337425
폴드 세트: 2 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 3 시작
macro-f1 score : 0.9922901558337425
폴드 세트: 4 시작
[32m[I 2023-04-13 13:54:08,370][0m Trial 1 finished with value: 0.9941404841551428 and parameters: {'criterion': 'gini', 'max_depth': 24}. Best is trial 1 with value: 0.9941404841551428.[0m
macro-f1 score : 0.9946035027418356
폴드 세트: 5 시작
macro-f1 score : 0.9961431963787019
폴드 세트: 1 시작
macro-f1 score : 0.990746804088489
폴드 세트: 2 시작
[32m[I 2023-04-13 13:54:08,653][0m Trial 2 finished with value: 0.9941403756734786 and parameters: {'criterion': 'log_loss', 'max_depth': 29}. Best is trial 1 with value: 0.9941404841551428.[0m
macro-f1 score : 0.9953754099876915
폴드 세트: 3 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 4 시작
macro-f1 score : 0.99306062581293
폴드 세트: 5 시작
macro-f1 score : 0.9969147677881807
폴드 세트: 1 시작
macro-f1 score : 0.9961464377374016
폴드 세트: 2 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 3 시작
[32m[I 2023-04-13 13:54:08,851][0m Trial 3 finished with value: 0.9941396892109434 and parameters: {'criterion': 'log_loss', 'max_depth': 20}. Best is trial 1 with value: 0.9941404841551428.[0m
[32m[I 2023-04-13 13:54:08,944][0m Trial 4 finished with value: 0.9941401366240677 and parameters: {'criterion': 'log_loss', 'max_depth': 23}. Best is trial 1 with value: 0.9941404841551428.[0m
macro-f1 score : 0.9946042706901019
폴드 세트: 4 시작
macro-f1 score : 0.993832124666994
폴드 세트: 5 시작
macro-f1 score : 0.9915113422701174
폴드 세트: 1 시작
macro-f1 score : 0.9915185426602477
폴드 세트: 2 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 3 시작
macro-f1 score : 0.9938330170777988
폴드 세트: 4 시작
macro-f1 score : 0.996145907635787
폴드 세트: 5 시작
macro-f1 score : 0.9938278057588139
폴드 세트: 1 시작
macro-f1 score :
[32m[I 2023-04-13 13:54:09,026][0m Trial 5 finished with value: 0.9941402235724436 and parameters: {'criterion': 'gini', 'max_depth': 22}. Best is trial 1 with value: 0.9941404841551428.[0m
[32m[I 2023-04-13 13:54:09,104][0m Trial 6 finished with value: 0.994139368965735 and parameters: {'criterion': 'log_loss', 'max_depth': 17}. Best is trial 1 with value: 0.9941404841551428.[0m
0.9946042706901019
폴드 세트: 2 시작
macro-f1 score : 0.9930616463823603
폴드 세트: 3 시작
macro-f1 score : 0.9915185426602477
폴드 세트: 4 시작
macro-f1 score : 0.9969169399917943
폴드 세트: 5 시작
macro-f1 score : 0.9945997181377133
폴드 세트: 1 시작
macro-f1 score : 0.9938330170777988
폴드 세트: 2 시작
macro-f1 score : 0.996917356704512
폴드 세트: 3 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 4 시작
macro-f1 score : 0.993832124666994
폴드 세트: 5 시작
macro-f1 score : 0.9907389363916785
폴드 세트: 1 시작
macro-f1 score : 0.9961464377374016
폴드 세트: 2 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 3 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 4 시작
[32m[I 2023-04-13 13:54:09,184][0m Trial 7 finished with value: 0.9941396861197674 and parameters: {'criterion': 'log_loss', 'max_depth': 18}. Best is trial 1 with value: 0.9941404841551428.[0m
[32m[I 2023-04-13 13:54:09,268][0m Trial 8 finished with value: 0.9941404295300813 and parameters: {'criterion': 'gini', 'max_depth': 12}. Best is trial 1 with value: 0.9941404841551428.[0m
[32m[I 2023-04-13 13:54:09,346][0m Trial 9 finished with value: 0.9941403027156713 and parameters: {'criterion': 'entropy', 'max_depth': 20}. Best is trial 1 with value: 0.9941404841551428.[0m
macro-f1 score : 0.9899733671272392
폴드 세트: 5 시작
macro-f1 score : 0.9938278057588139
폴드 세트: 1 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 2 시작
macro-f1 score : 0.9938330170777988
폴드 세트: 3 시작
macro-f1 score : 0.9938330170777988
폴드 세트: 4 시작
macro-f1 score : 0.993832124666994
폴드 세트: 5 시작
macro-f1 score : 0.9945997181377133
폴드 세트: 1 시작
macro-f1 score : 0.990746804088489
폴드 세트: 2 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 3 시작
macro-f1 score : 0.9938330170777988
폴드 세트: 4 시작
macro-f1 score : 0.996145907635787
폴드 세트: 5 시작
macro-f1 score : 0.9953715140861799
폴드 세트: 1 시작
macro-f1 score : 0.996917356704512
[32m[I 2023-04-13 13:54:09,437][0m Trial 10 finished with value: 0.9941397625237048 and parameters: {'criterion': 'entropy', 'max_depth': 4}. Best is trial 1 with value: 0.9941404841551428.[0m
[32m[I 2023-04-13 13:54:09,529][0m Trial 11 finished with value: 0.994139257416301 and parameters: {'criterion': 'gini', 'max_depth': 12}. Best is trial 1 with value: 0.9941404841551428.[0m
폴드 세트: 2 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 3 시작
macro-f1 score : 0.9938330170777988
폴드 세트: 4 시작
macro-f1 score : 0.9915172546751494
폴드 세트: 5 시작
macro-f1 score : 0.9930557741733722
폴드 세트: 1 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 2 시작
macro-f1 score : 0.9976881696527808
폴드 세트: 3 시작
macro-f1 score : 0.9930616463823603
폴드 세트: 4 시작
macro-f1 score : 0.993832124666994
폴드 세트: 5 시작
macro-f1 score : 0.9907389363916785
폴드 세트: 1 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 2 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 3 시작
[32m[I 2023-04-13 13:54:09,615][0m Trial 12 finished with value: 0.9941402258654672 and parameters: {'criterion': 'gini', 'max_depth': 11}. Best is trial 1 with value: 0.9941404841551428.[0m
[32m[I 2023-04-13 13:54:09,698][0m Trial 13 finished with value: 0.99414016387632 and parameters: {'criterion': 'gini', 'max_depth': 12}. Best is trial 1 with value: 0.9941404841551428.[0m
[32m[I 2023-04-13 13:54:09,784][0m Trial 14 finished with value: 0.9941401589374108 and parameters: {'criterion': 'gini', 'max_depth': 30}. Best is trial 1 with value: 0.9941404841551428.[0m
macro-f1 score : 0.9938330170777988
폴드 세트: 4 시작
macro-f1 score : 0.99306062581293
폴드 세트: 5 시작
macro-f1 score : 0.9938278057588139
폴드 세트: 1 시작
macro-f1 score : 0.990746804088489
폴드 세트: 2 시작
macro-f1 score : 0.9961464377374016
폴드 세트: 3 시작
macro-f1 score : 0.9922901558337425
폴드 세트: 4 시작
macro-f1 score : 0.996145907635787
폴드 세트: 5 시작
macro-f1 score : 0.9953715140861799
폴드 세트: 1 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 2 시작
macro-f1 score : 0.9961464377374016
폴드 세트: 3 시작
macro-f1 score : 0.9922901558337425
폴드 세트: 4 시작
macro-f1 score : 0.993832124666994
폴드 세트: 5 시작
macro-f1 score : 0.9938278057588139
[32m[I 2023-04-13 13:54:09,870][0m Trial 15 finished with value: 0.9941402078743946 and parameters: {'criterion': 'gini', 'max_depth': 6}. Best is trial 1 with value: 0.9941404841551428.[0m
[32m[I 2023-04-13 13:54:09,961][0m Trial 16 finished with value: 0.9941404810181729 and parameters: {'criterion': 'gini', 'max_depth': 26}. Best is trial 1 with value: 0.9941404841551428.[0m
폴드 세트: 1 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 2 시작
macro-f1 score : 0.9899749373433584
폴드 세트: 3 시작
macro-f1 score : 0.9961464377374016
폴드 세트: 4 시작
macro-f1 score : 0.99306062581293
폴드 세트: 5 시작
macro-f1 score : 0.9969147677881807
폴드 세트: 1 시작
macro-f1 score : 0.9922901558337425
폴드 세트: 2 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 3 시작
macro-f1 score : 0.9938330170777988
폴드 세트: 4 시작
macro-f1 score : 0.99306062581293
폴드 세트: 5 시작
macro-f1 score : 0.9961431963787019
폴드 세트: 1 시작
macro-f1 score : 0.9930616463823603
폴드 세트: 2 시작
macro-f1 score : 0.9930616463823603
[32m[I 2023-04-13 13:54:10,055][0m Trial 17 finished with value: 0.9941400534967688 and parameters: {'criterion': 'entropy', 'max_depth': 26}. Best is trial 1 with value: 0.9941404841551428.[0m
[32m[I 2023-04-13 13:54:10,144][0m Trial 18 finished with value: 0.994140178983136 and parameters: {'criterion': 'gini', 'max_depth': 27}. Best is trial 1 with value: 0.9941404841551428.[0m
폴드 세트: 3 시작
macro-f1 score : 0.9961464377374016
폴드 세트: 4 시작
macro-f1 score : 0.9953747628083491
폴드 세트: 5 시작
macro-f1 score : 0.9930557741733722
폴드 세트: 1 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 2 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 3 시작
macro-f1 score : 0.9899749373433584
폴드 세트: 4 시작
macro-f1 score : 0.9953747628083491
폴드 세트: 5 시작
macro-f1 score : 0.9953715140861799
폴드 세트: 1 시작
macro-f1 score : 0.9922901558337425
폴드 세트: 2 시작
macro-f1 score : 0.9930616463823603
폴드 세트: 3 시작
macro-f1 score : 0.9915185426602477
폴드 세트: 4 시작
[32m[I 2023-04-13 13:54:10,233][0m Trial 19 finished with value: 0.9941404967196202 and parameters: {'criterion': 'gini', 'max_depth': 27}. Best is trial 19 with value: 0.9941404967196202.[0m
macro-f1 score : 0.996145907635787
폴드 세트: 5 시작
macro-f1 score : 0.9976862310859633
cbrm_trial = optuna_cbrm.best_trial
cbrm_trial_params = cbrm_trial.params
print('Best Trial: score {},\nparams {}'.format(cbrm_trial.value, cbrm_trial_params))
Best Trial: score 0.9941404967196202,
params {'criterion': 'gini', 'max_depth': 27}
model2=DecisionTreeClassifier(**cbrm_trial_params)
model2.fit(train_df,y)
DecisionTreeClassifier(max_depth=27)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
DecisionTreeClassifier(max_depth=27)
y_test_pred=model2.predict(test_df)
y_test_proba=model2.predict_proba(test_df)
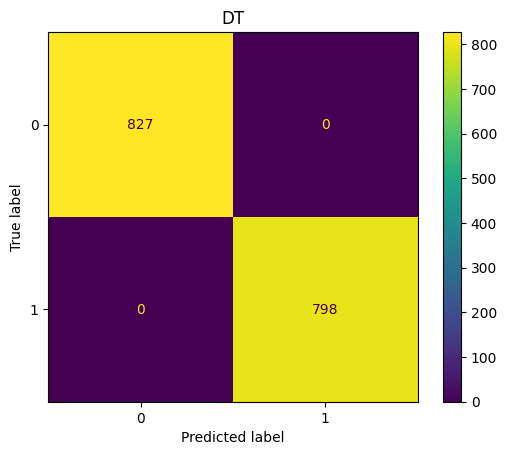
accuracy_score(y_test,y_test_pred)
1.0
f1_score(y_test,y_test_pred,average='macro')
1.0
DT Confusion Matrix
show_confusion(y_test_pred,y_test,"DT")
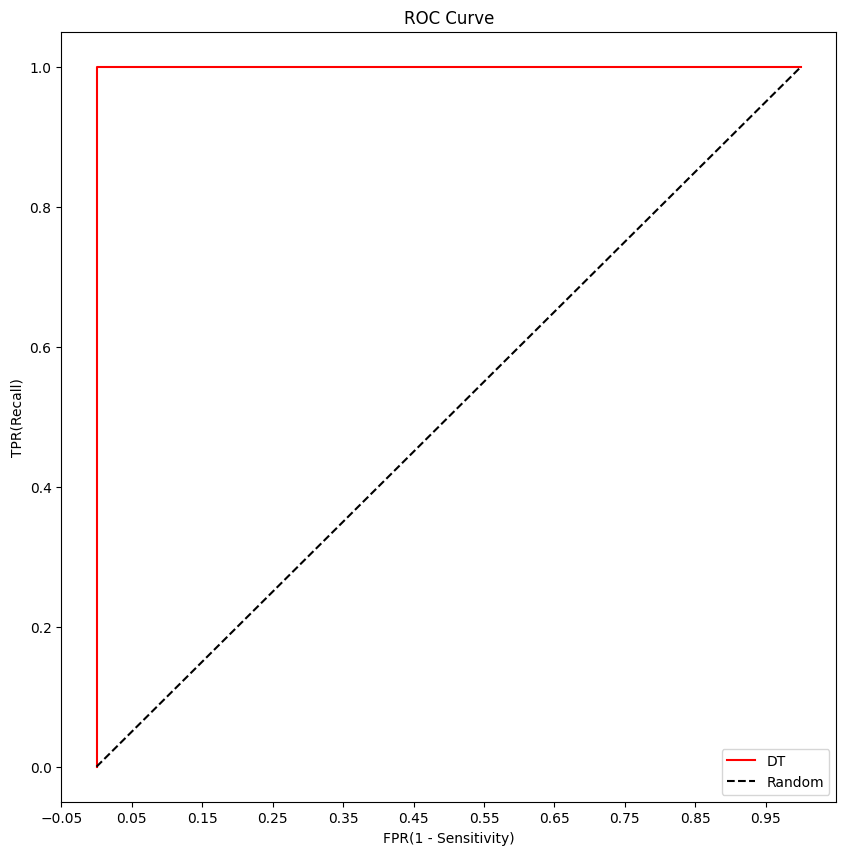
DT Roc Curve
plt.figure(figsize=(10,10))
fprs, tprs, thresholds = roc_curve(y_test, y_test_proba[:,1])
# ROC 곡선 그래프 곡선으로 그림
plt.plot(fprs, tprs, label='DT',color='red')
plt.plot([0,1],[0,1], 'k--', label='Random')
# FPR X 축의 Scale을 0.1 단위로 변경, X, Y 축 명 설정 등
start,end = plt.xlim()
plt.xticks(np.round(np.arange(start, end, 0.1), 2))
#plt.xlim(0, 1); plt.ylim(0, 1)
plt.xlabel('FPR(1 - Sensitivity)'); plt.ylabel('TPR(Recall)')
plt.title('ROC Curve')
plt.legend()
<matplotlib.legend.Legend at 0x7f3f340c3b50>
MLP
파라미터 튜닝
# 데이터 분리에서 x_train , x_valid , y_train , y_valid
#분리해놓은 것을 통해 최적 파라미터를 탐색한다.
sampler = TPESampler(seed=10)
# define function
def objective(trial):
cbrm_param = {
"learning_rate": trial.suggest_categorical("learning_rate",['constant','invscaling','adaptive']),
"alpha": trial.suggest_categorical("alpha",[.3,.1,.01,.001,.0001]),
"activation": trial.suggest_categorical("activation",['logistic','relu','tanh'])
}
n_folds=5
kf = StratifiedKFold(n_splits=n_folds, shuffle=True)
# 추후 메타 모델이 사용할 학습 데이터 반환을 위한 넘파이 배열 초기화
model_cbrm=MLPClassifier(**cbrm_param)
m_f1 =0.0
for folder_counter, (train_index, valid_index) in enumerate(kf.split(train,y)):
# 입력된 학습 데이터에서 기반 모델이 학습/예측할 폴드 데이터 세트 추출
print('\t 폴드 세트: ',folder_counter+1,' 시작')
X_tr = train.loc[train_index]
y_tr = y.loc[train_index]
X_te = train.loc[valid_index]
y_te = y.loc[valid_index]
model_cbrm = model_cbrm.fit(X_tr, y_tr)
pred=model_cbrm.predict(X_te)
print("macro-f1 score : ",f1_score(y_te,pred,average='macro'))
m_f1+=f1_score(y_te,pred,average='macro')
# 평가지표 원하는 평가 지표가 있을 시 바꾸어 준다.
m_f1 = m_f1 / n_folds
return m_f1
optuna_cbrm = optuna.create_study(direction='maximize', sampler=sampler)
optuna_cbrm.optimize(objective, n_trials=4)
[32m[I 2023-04-13 13:55:12,348][0m A new study created in memory with name: no-name-15a4683e-0212-4c96-96bc-df64bbd267ef[0m
폴드 세트: 1 시작
macro-f1 score : 0.9930616463823603
폴드 세트: 2 시작
macro-f1 score : 0.9938330170777988
폴드 세트: 3 시작
macro-f1 score : 0.9922901558337425
폴드 세트: 4 시작
macro-f1 score : 0.993832124666994
폴드 세트: 5 시작
[32m[I 2023-04-13 13:55:26,491][0m Trial 0 finished with value: 0.9941406350093718 and parameters: {'learning_rate': 'constant', 'alpha': 0.0001, 'activation': 'tanh'}. Best is trial 0 with value: 0.9941406350093718.[0m
macro-f1 score : 0.9976862310859633
폴드 세트: 1 시작
macro-f1 score : 0.9930616463823603
폴드 세트: 2 시작
macro-f1 score : 0.9922901558337425
폴드 세트: 3 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 4 시작
macro-f1 score : 0.99306062581293
폴드 세트: 5 시작
[32m[I 2023-04-13 13:55:44,619][0m Trial 1 finished with value: 0.9941405211609811 and parameters: {'learning_rate': 'constant', 'alpha': 0.0001, 'activation': 'logistic'}. Best is trial 0 with value: 0.9941406350093718.[0m
macro-f1 score : 0.9969147677881807
폴드 세트: 1 시작
macro-f1 score : 0.9922901558337425
폴드 세트: 2 시작
macro-f1 score : 0.996917356704512
폴드 세트: 3 시작
macro-f1 score : 0.9953754099876915
폴드 세트: 4 시작
macro-f1 score : 0.9915172546751494
폴드 세트: 5 시작
[32m[I 2023-04-13 13:55:59,304][0m Trial 2 finished with value: 0.9941399790677619 and parameters: {'learning_rate': 'invscaling', 'alpha': 0.001, 'activation': 'tanh'}. Best is trial 0 with value: 0.9941406350093718.[0m
macro-f1 score : 0.9945997181377133
폴드 세트: 1 시작
macro-f1 score : 0.9938330170777988
폴드 세트: 2 시작
macro-f1 score : 0.9922901558337425
폴드 세트: 3 시작
macro-f1 score : 0.9946042706901019
폴드 세트: 4 시작
macro-f1 score : 0.9953747628083491
폴드 세트: 5 시작
[32m[I 2023-04-13 13:56:23,298][0m Trial 3 finished with value: 0.9941403849095412 and parameters: {'learning_rate': 'constant', 'alpha': 0.1, 'activation': 'tanh'}. Best is trial 0 with value: 0.9941406350093718.[0m
macro-f1 score : 0.9945997181377133
cbrm_trial = optuna_cbrm.best_trial
cbrm_trial_params = cbrm_trial.params
print('Best Trial: score {},\nparams {}'.format(cbrm_trial.value, cbrm_trial_params))
Best Trial: score 0.9941406350093718,
params {'learning_rate': 'constant', 'alpha': 0.0001, 'activation': 'tanh'}
mlp=MLPClassifier(**cbrm_trial_params)
mlp.fit(train_df,y)
MLPClassifier(activation='tanh')In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
MLPClassifier(activation='tanh')
y_test_pred=mlp.predict(test_df)
y_test_proba=mlp.predict_proba(test_df)
accuracy_score(y_test,y_test_pred)
1.0
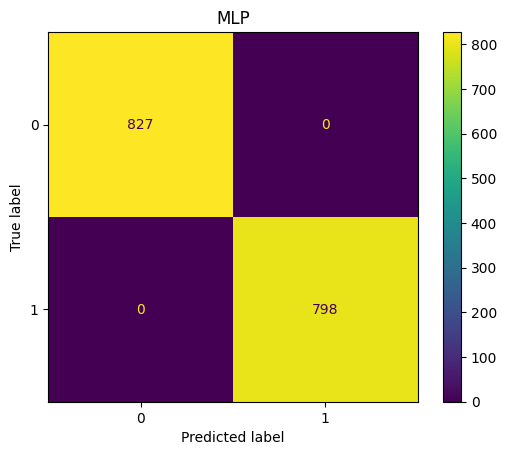
MLP Confusion Matrix
show_confusion(y_test_pred,y_test,"MLP")
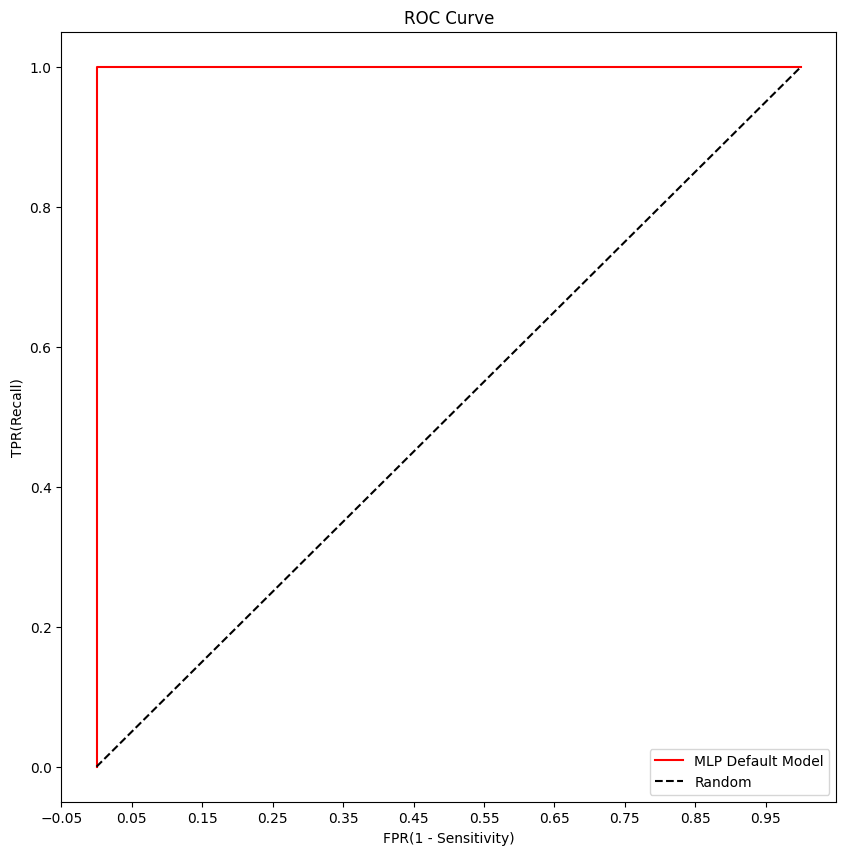
MLP Roc Curve
plt.figure(figsize=(10,10))
fprs, tprs, thresholds = roc_curve(y_test, y_test_proba[:,1])
# ROC 곡선 그래프 곡선으로 그림
plt.plot(fprs, tprs, label='MLP Default Model',color='red')
plt.plot([0,1],[0,1], 'k--', label='Random')
# FPR X 축의 Scale을 0.1 단위로 변경, X, Y 축 명 설정 등
start,end = plt.xlim()
plt.xticks(np.round(np.arange(start, end, 0.1), 2))
#plt.xlim(0, 1); plt.ylim(0, 1)
plt.xlabel('FPR(1 - Sensitivity)'); plt.ylabel('TPR(Recall)')
plt.title('ROC Curve')
plt.legend()
<matplotlib.legend.Legend at 0x7f3f3460fd00>
DNN
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.utils import to_categorical
# train : train, y / test : test / y_test
y_test
0 0
1 1
2 1
3 0
4 0
..
1620 1
1621 0
1622 1
1623 1
1624 1
Name: class, Length: 1625, dtype: int64
y_Train=to_categorical(y)
y_Test = to_categorical(y_test)
train.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 6499 entries, 0 to 6498
Data columns (total 18 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 odor_0 6499 non-null float64
1 odor_1 6499 non-null float64
2 odor_2 6499 non-null float64
3 odor_3 6499 non-null float64
4 odor_4 6499 non-null float64
5 odor_5 6499 non-null float64
6 odor_6 6499 non-null float64
7 odor_7 6499 non-null float64
8 odor_8 6499 non-null float64
9 spore-print-color_0 6499 non-null float64
10 spore-print-color_1 6499 non-null float64
11 spore-print-color_2 6499 non-null float64
12 spore-print-color_3 6499 non-null float64
13 spore-print-color_4 6499 non-null float64
14 spore-print-color_5 6499 non-null float64
15 spore-print-color_6 6499 non-null float64
16 spore-print-color_7 6499 non-null float64
17 spore-print-color_8 6499 non-null float64
dtypes: float64(18)
memory usage: 914.0 KB
model = keras.Sequential([keras.layers.Dense(5,input_shape=(18,)),
keras.layers.Dense(3,activation='relu'),
keras.layers.Dense(2,activation='softmax')])
model.summary()
Model: "sequential_6"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_18 (Dense) (None, 5) 95
dense_19 (Dense) (None, 3) 18
dense_20 (Dense) (None, 2) 8
=================================================================
Total params: 121
Trainable params: 121
Non-trainable params: 0
_________________________________________________________________
train.shape
(6499, 9)
model.compile(optimizer='adam',loss='binary_crossentropy',metrics=['accuracy'])
history = model.fit(train,y_Train,validation_split=0.1,epochs=25)
Epoch 1/25
183/183 [==============================] - 2s 5ms/step - loss: 0.6181 - accuracy: 0.7812 - val_loss: 0.5082 - val_accuracy: 0.9446
Epoch 2/25
183/183 [==============================] - 0s 2ms/step - loss: 0.4422 - accuracy: 0.9085 - val_loss: 0.3348 - val_accuracy: 0.9446
Epoch 3/25
183/183 [==============================] - 0s 2ms/step - loss: 0.3134 - accuracy: 0.9598 - val_loss: 0.2488 - val_accuracy: 0.9892
Epoch 4/25
183/183 [==============================] - 0s 2ms/step - loss: 0.2489 - accuracy: 0.9846 - val_loss: 0.2075 - val_accuracy: 0.9892
Epoch 5/25
183/183 [==============================] - 0s 2ms/step - loss: 0.2144 - accuracy: 0.9846 - val_loss: 0.1802 - val_accuracy: 0.9892
Epoch 6/25
183/183 [==============================] - 0s 2ms/step - loss: 0.1888 - accuracy: 0.9846 - val_loss: 0.1589 - val_accuracy: 0.9892
Epoch 7/25
183/183 [==============================] - 0s 2ms/step - loss: 0.1680 - accuracy: 0.9846 - val_loss: 0.1410 - val_accuracy: 0.9892
Epoch 8/25
183/183 [==============================] - 0s 2ms/step - loss: 0.1504 - accuracy: 0.9846 - val_loss: 0.1260 - val_accuracy: 0.9892
Epoch 9/25
183/183 [==============================] - 0s 2ms/step - loss: 0.1353 - accuracy: 0.9915 - val_loss: 0.1129 - val_accuracy: 0.9969
Epoch 10/25
183/183 [==============================] - 0s 2ms/step - loss: 0.1221 - accuracy: 0.9938 - val_loss: 0.1011 - val_accuracy: 0.9969
Epoch 11/25
183/183 [==============================] - 0s 2ms/step - loss: 0.1099 - accuracy: 0.9938 - val_loss: 0.0903 - val_accuracy: 0.9969
Epoch 12/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0986 - accuracy: 0.9938 - val_loss: 0.0807 - val_accuracy: 0.9969
Epoch 13/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0892 - accuracy: 0.9938 - val_loss: 0.0728 - val_accuracy: 0.9969
Epoch 14/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0817 - accuracy: 0.9938 - val_loss: 0.0665 - val_accuracy: 0.9969
Epoch 15/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0756 - accuracy: 0.9938 - val_loss: 0.0610 - val_accuracy: 0.9969
Epoch 16/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0704 - accuracy: 0.9938 - val_loss: 0.0564 - val_accuracy: 0.9969
Epoch 17/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0658 - accuracy: 0.9938 - val_loss: 0.0523 - val_accuracy: 0.9969
Epoch 18/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0619 - accuracy: 0.9938 - val_loss: 0.0487 - val_accuracy: 0.9969
Epoch 19/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0584 - accuracy: 0.9938 - val_loss: 0.0455 - val_accuracy: 0.9969
Epoch 20/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0554 - accuracy: 0.9938 - val_loss: 0.0427 - val_accuracy: 0.9969
Epoch 21/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0527 - accuracy: 0.9938 - val_loss: 0.0402 - val_accuracy: 0.9969
Epoch 22/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0504 - accuracy: 0.9938 - val_loss: 0.0379 - val_accuracy: 0.9969
Epoch 23/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0483 - accuracy: 0.9938 - val_loss: 0.0360 - val_accuracy: 0.9969
Epoch 24/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0465 - accuracy: 0.9938 - val_loss: 0.0342 - val_accuracy: 0.9969
Epoch 25/25
183/183 [==============================] - 0s 2ms/step - loss: 0.0449 - accuracy: 0.9938 - val_loss: 0.0327 - val_accuracy: 0.9969
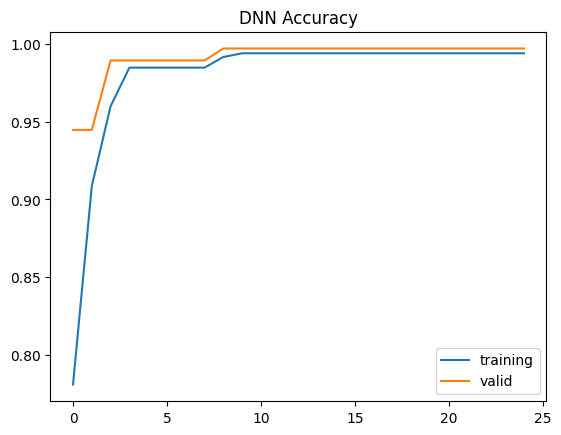
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
plt.title("DNN Accuracy")
plt.legend(['training','valid'],loc='best')
plt.show()
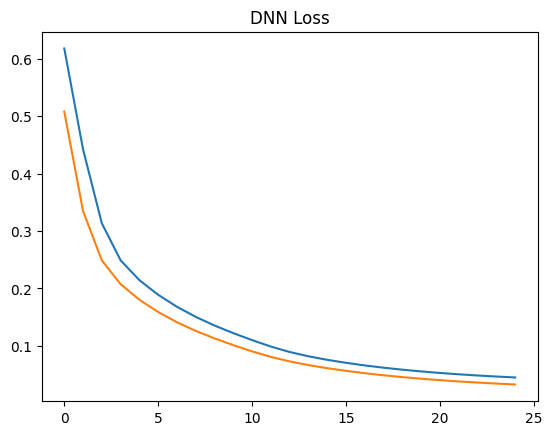
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title("DNN Loss")
plt.show()
y_test
0 0
1 1
2 1
3 0
4 0
..
1620 1
1621 0
1622 1
1623 1
1624 1
Name: class, Length: 1625, dtype: int64
y_pred=model.predict(test)
y_pred_labels = []
for i in y_pred :
if (np.argmax(i)>0):
y_pred_labels.append(1)
else :
y_pred_labels.append(0)
51/51 [==============================] - 0s 1ms/step
accuracy_score(y_test,y_pred_labels)
0.9938461538461538
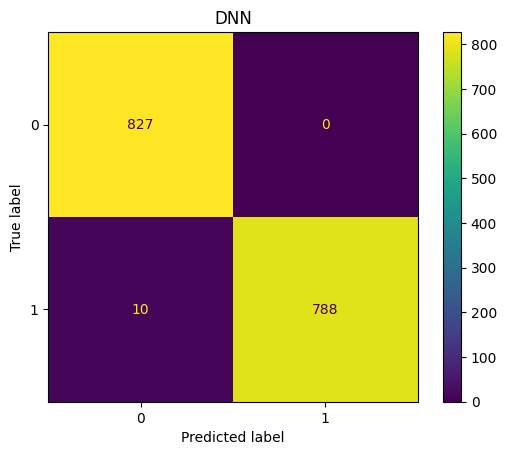
show_confusion(y_pred_labels , y_test,'DNN')

댓글남기기